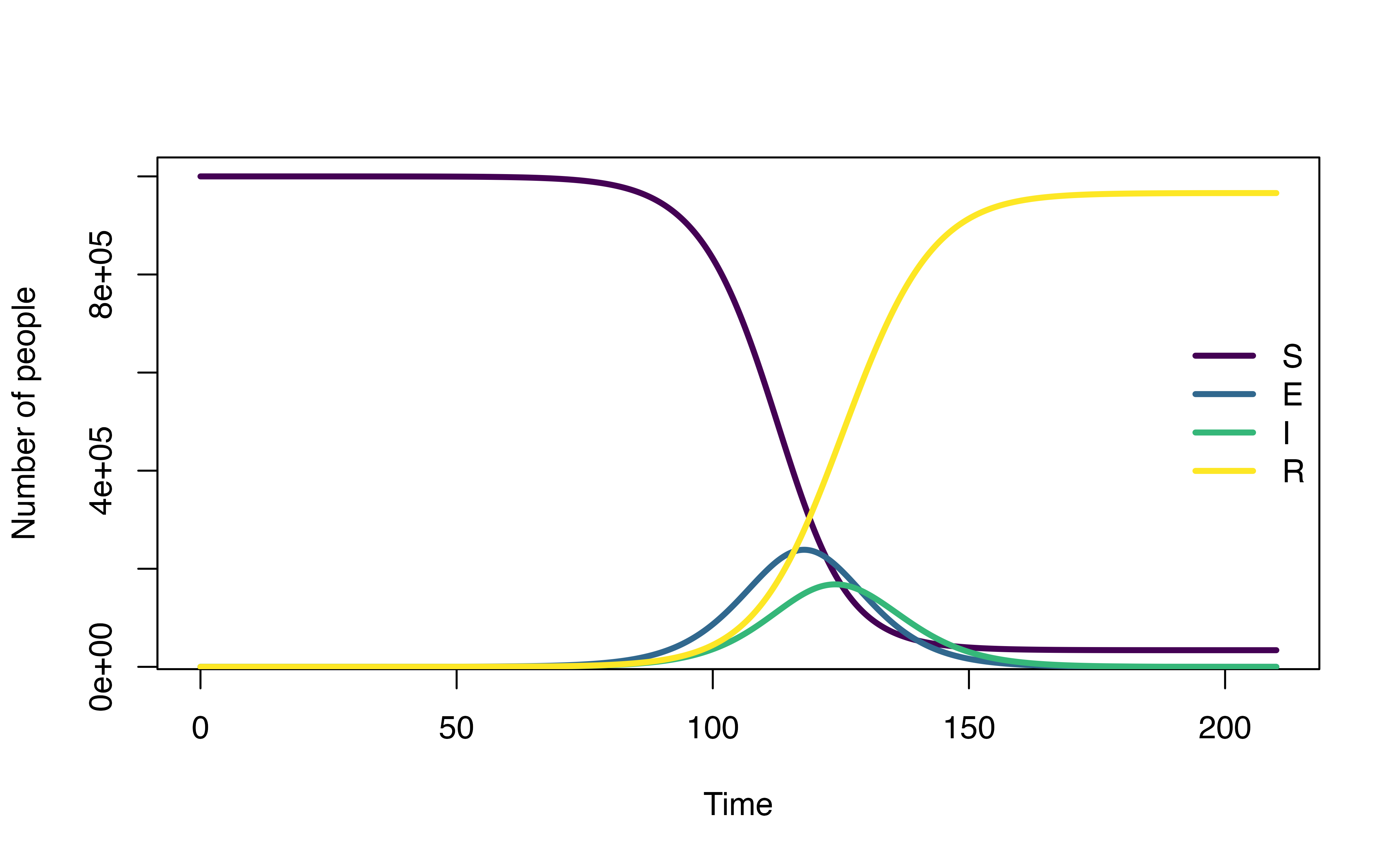Comparison between deSolve and denim
## Warning: package 'deSolve' was built under R version 4.3.1
# --- Timestep definition
simulationDuration <- 300
timestep <- 0.01 # small timestep required for comparison
# --- Transition def for denim
transitions <- list(
# note that timeStep uses the timeStep parameter in denim::sim()
"S -> I" = "beta * S * (I/N) * timeStep",
"I -> R" = d_gamma(rate = 1/3, shape = 2)
)
parameters <- c(beta = 0.2, rate = 1/3, shape=2, N=1000)
initialValues <- c(S = 999, I = 1, I1 = 1, I2=0, R=0)
# --- Transition def for deSolve
transition_func <- function(t, state, param){
with(as.list( c(state, param) ), {
dS = - beta * S * I/N
# apply linear chain trick
dI1 = beta * S * I/N - rate*I1
dI2 = rate*I1 - rate*I2
dI = dI1 + dI2
dR = rate*I2
list(c(dS, dI, dI1, dI2, dR))
})
}Run simulation with denim
denim_runtime <- bench::mark(
sim(transitions = transitions, initialValues = initialValues, parameters, simulationDuration = simulationDuration, timeStep = timestep),
iterations = 1
)
denim_runtime <- as.numeric(denim_runtime$total_time)
mod <- sim(transitions = transitions, initialValues = initialValues, parameters, simulationDuration = simulationDuration, timeStep = timestep)
# --- show output
head(mod[mod$Time %in% 1:simulationDuration,])## Time S I R
## 101 1 998.7825 1.169597 0.04786644
## 201 2 998.5358 1.296591 0.16756473
## 301 3 998.2665 1.399083 0.33440701
## 401 4 997.9783 1.487980 0.53374794
## 501 5 997.6732 1.569868 0.75697598
## 601 6 997.3521 1.648745 0.99910771Run simulation with deSolve
times <- seq(0, simulationDuration, timestep)
desolve_runtime <- bench::mark(
ode(y = initialValues, times = times, parms = parameters, func = transition_func),
iterations = 1
)## Warning: Some expressions had a GC in every iteration; so filtering is
## disabled.
desolve_runtime <- as.numeric(desolve_runtime$total_time)
ode_mod <- ode(y = initialValues, times = times, parms = parameters, func = transition_func)
# --- show output
ode_mod <- as.data.frame(ode_mod)
head(ode_mod[ode_mod$time %in% 1:simulationDuration, c("time", "S", "I", "R")])## time S I R
## 101 1 998.7824 1.169731 0.0479167
## 201 2 998.5355 1.296752 0.1677463
## 301 3 998.2660 1.399189 0.3347737
## 401 4 997.9777 1.487967 0.5343309
## 501 5 997.6725 1.569688 0.7577887
## 601 6 997.3515 1.648357 1.0001499Execution time comparison
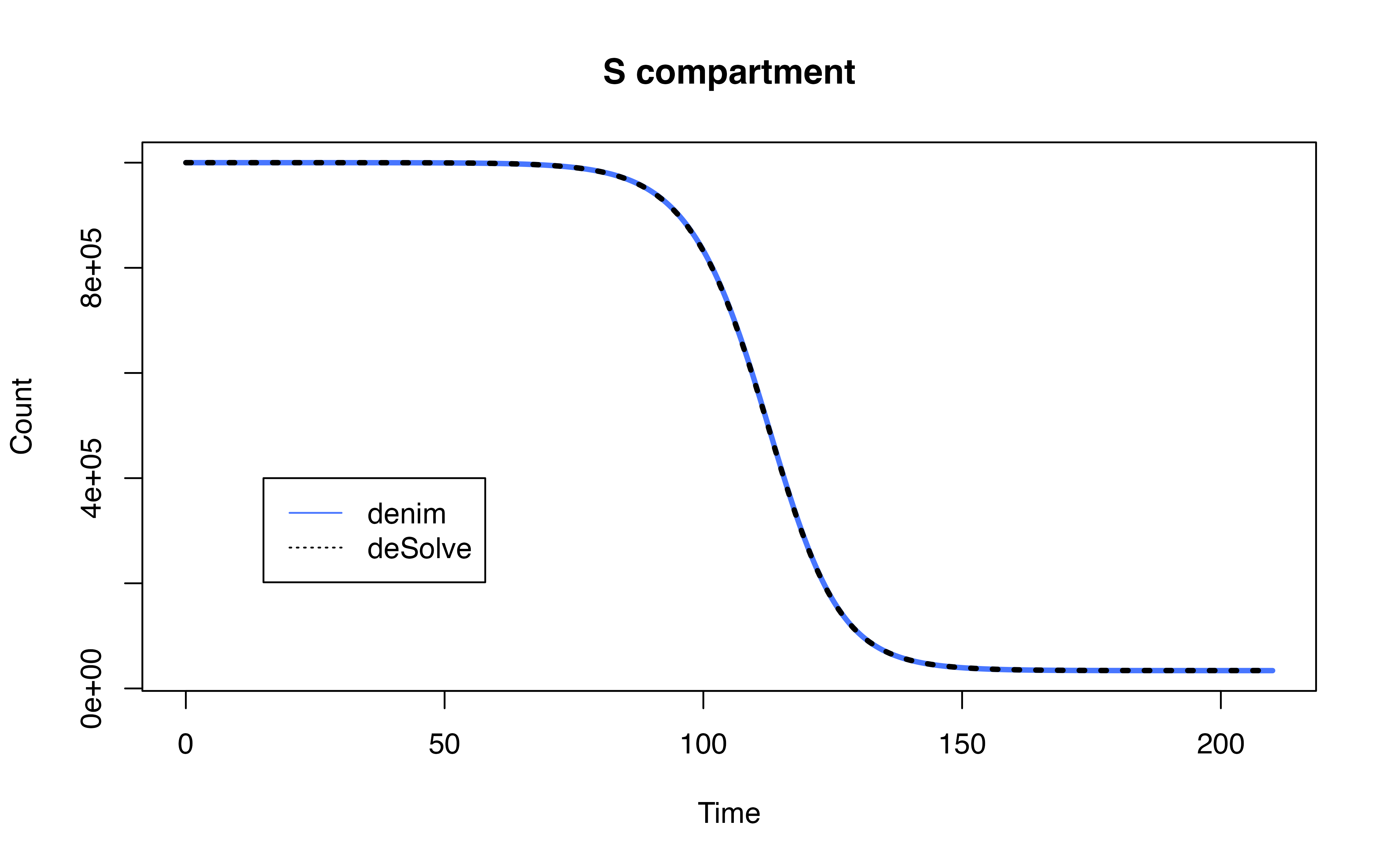
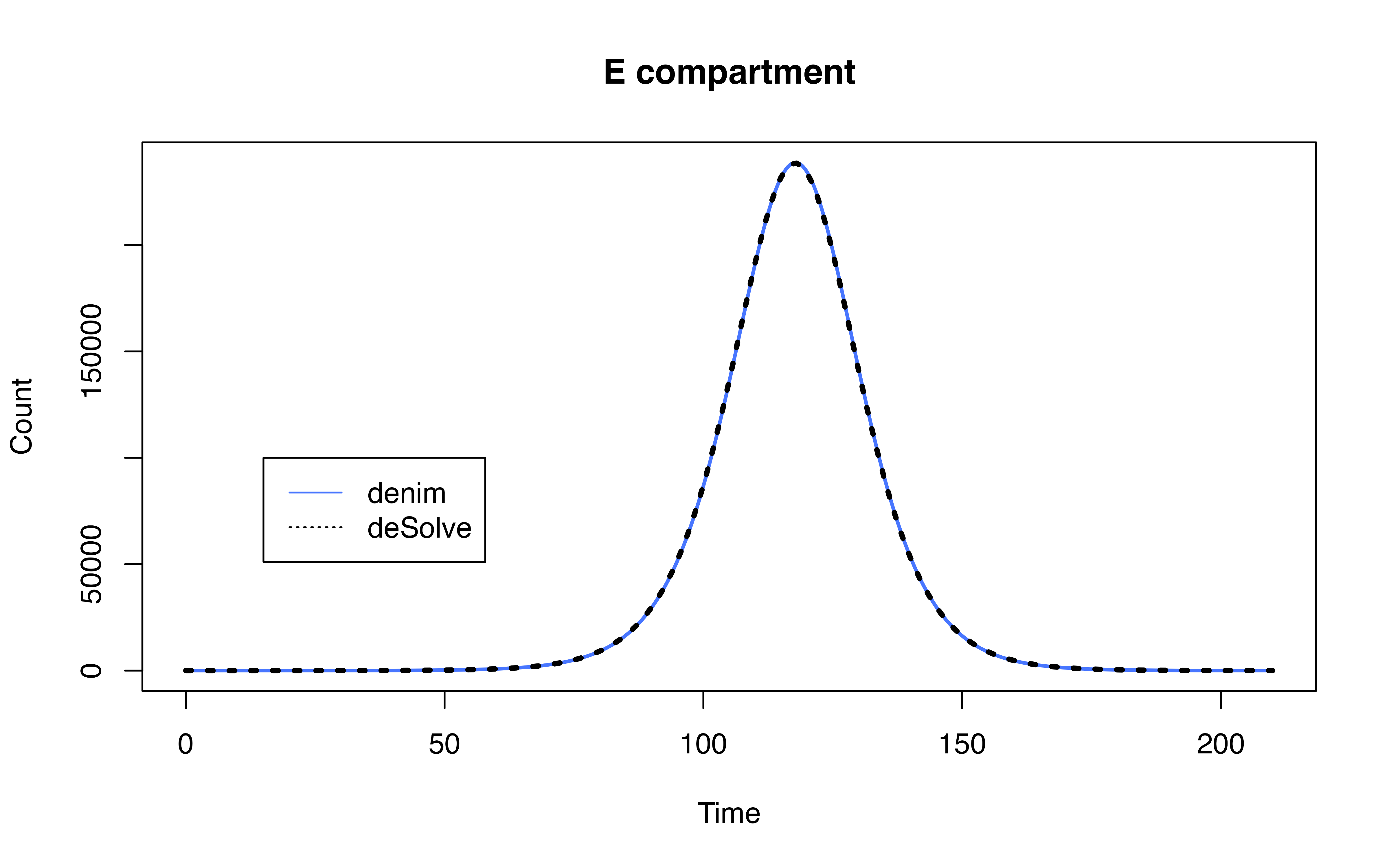
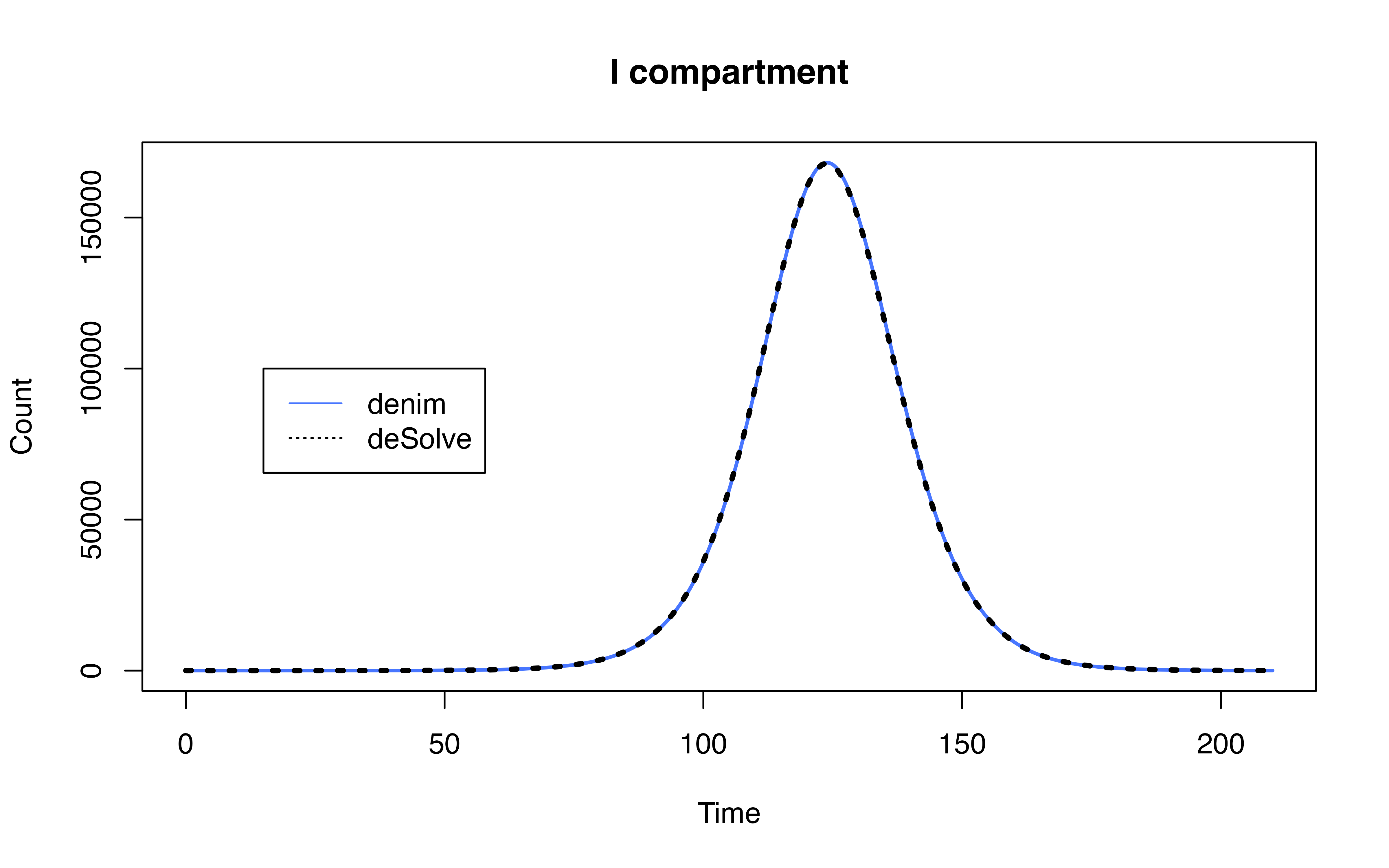
denim takes approximately 71.18 times as long as
deSolve to compute the result with the given specifications
.
This significant difference can be attributed to the difference in
approaches: deSolve solves a system of ODEs while
denim iterates through each timestep and updates the
population in each compartment
While the approach in denim allow more flexibility in
types of dwell time distributions, the computation time scales up
exponentially as timestep grows smaller.
Plot the result
# increase timestep before plotting
mod <- mod[mod$Time %in% seq(0, simulationDuration, 0.2),]
ode_mod <- ode_mod[ode_mod$time %in% seq(0, simulationDuration, 0.2),]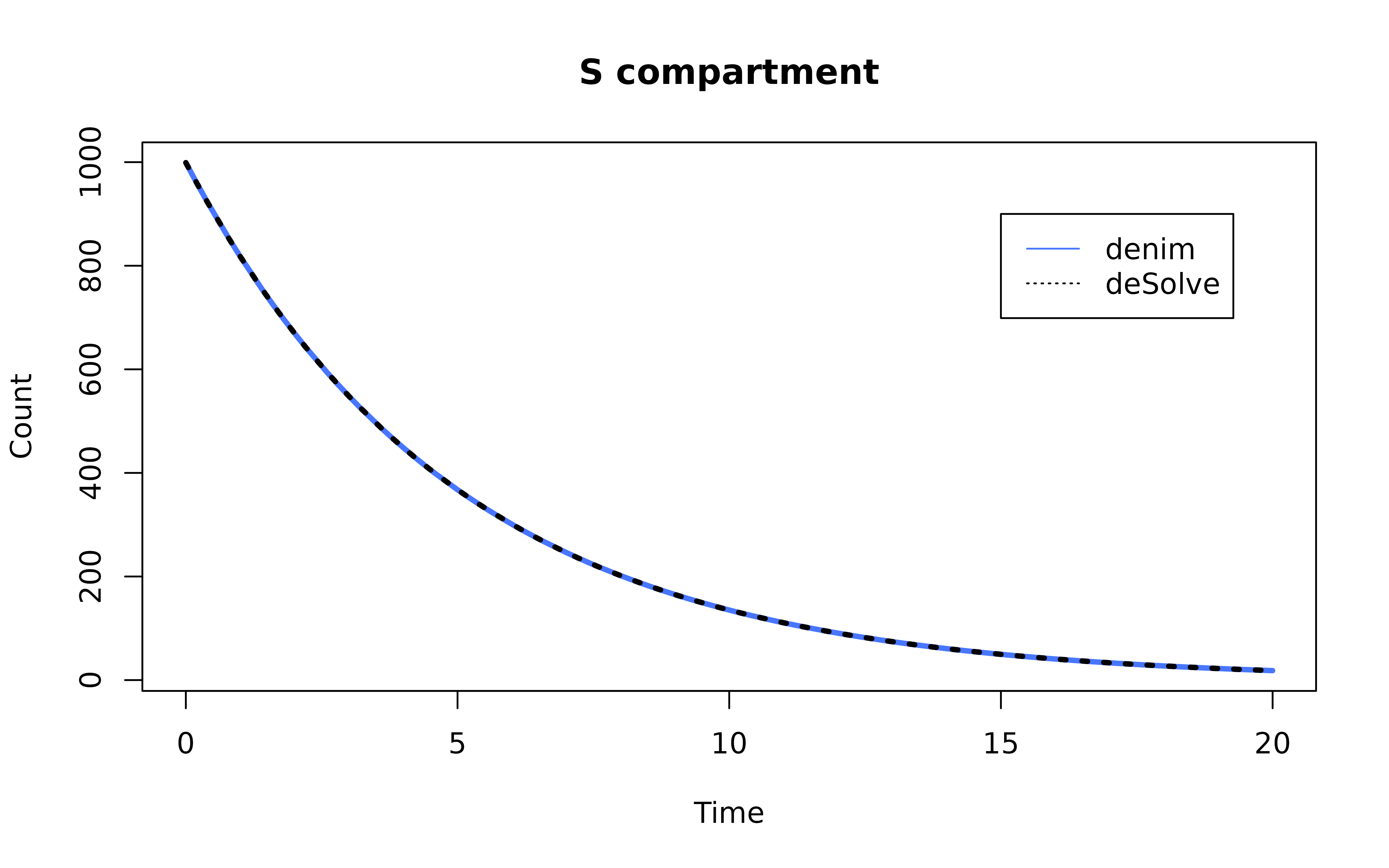
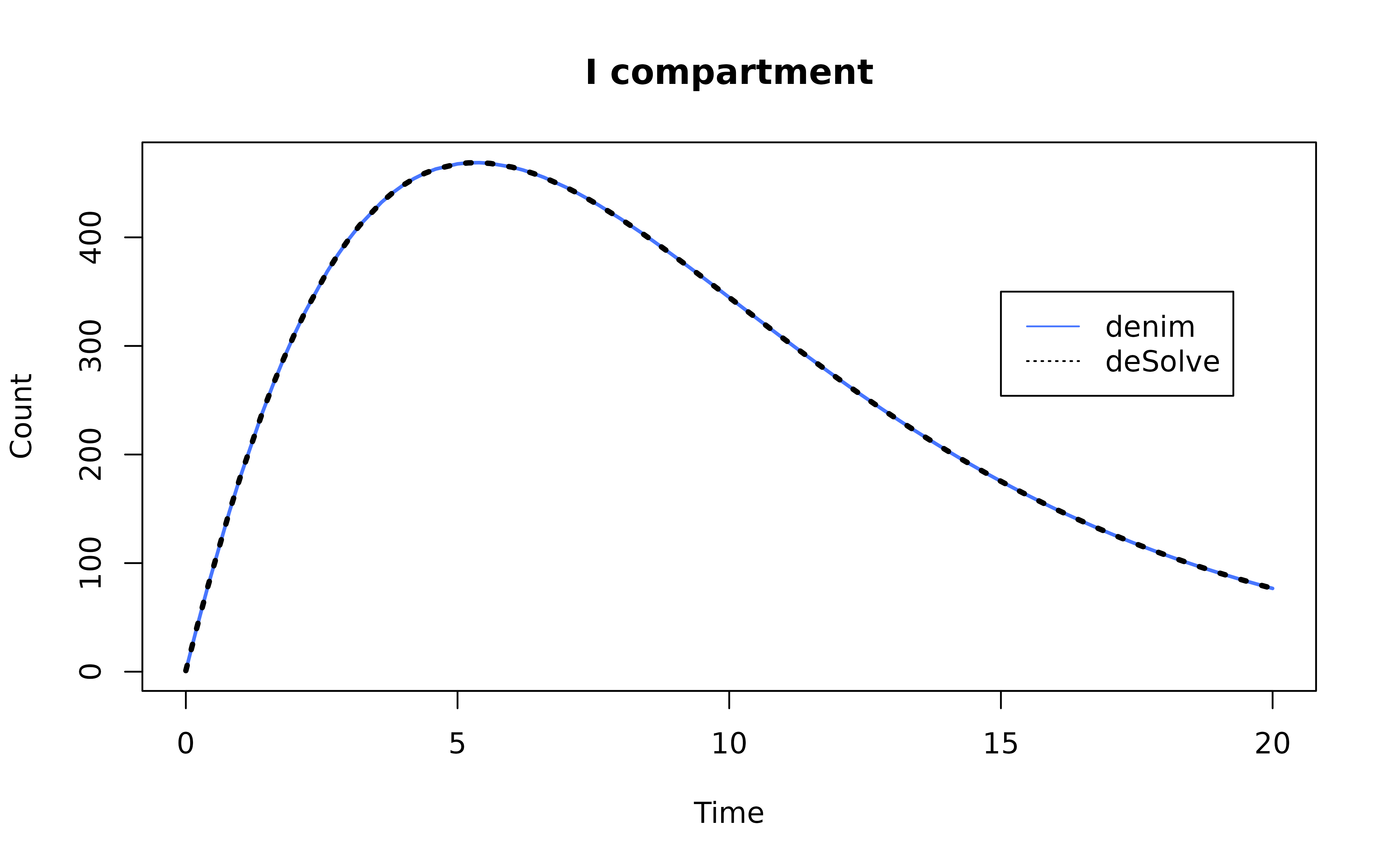
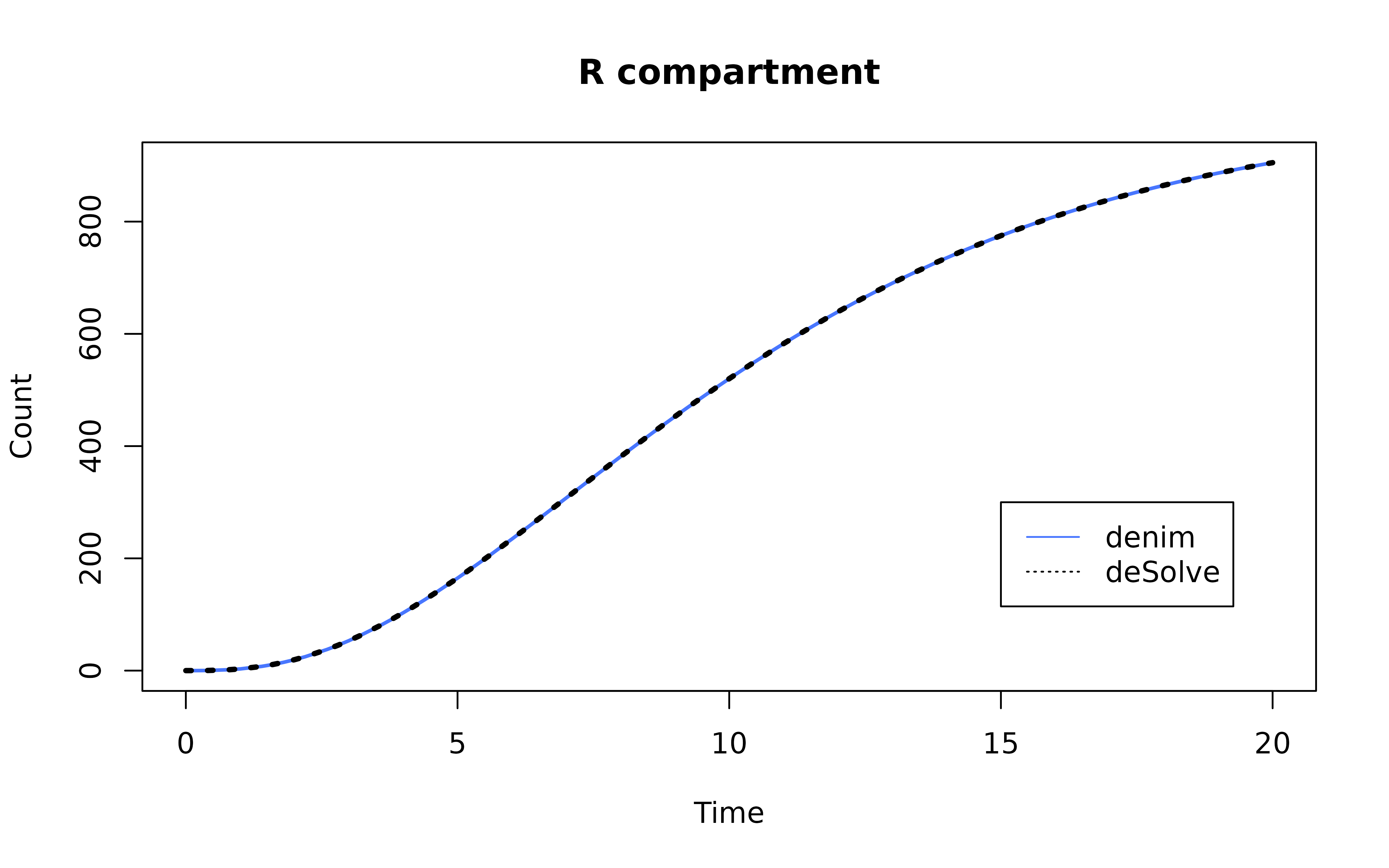
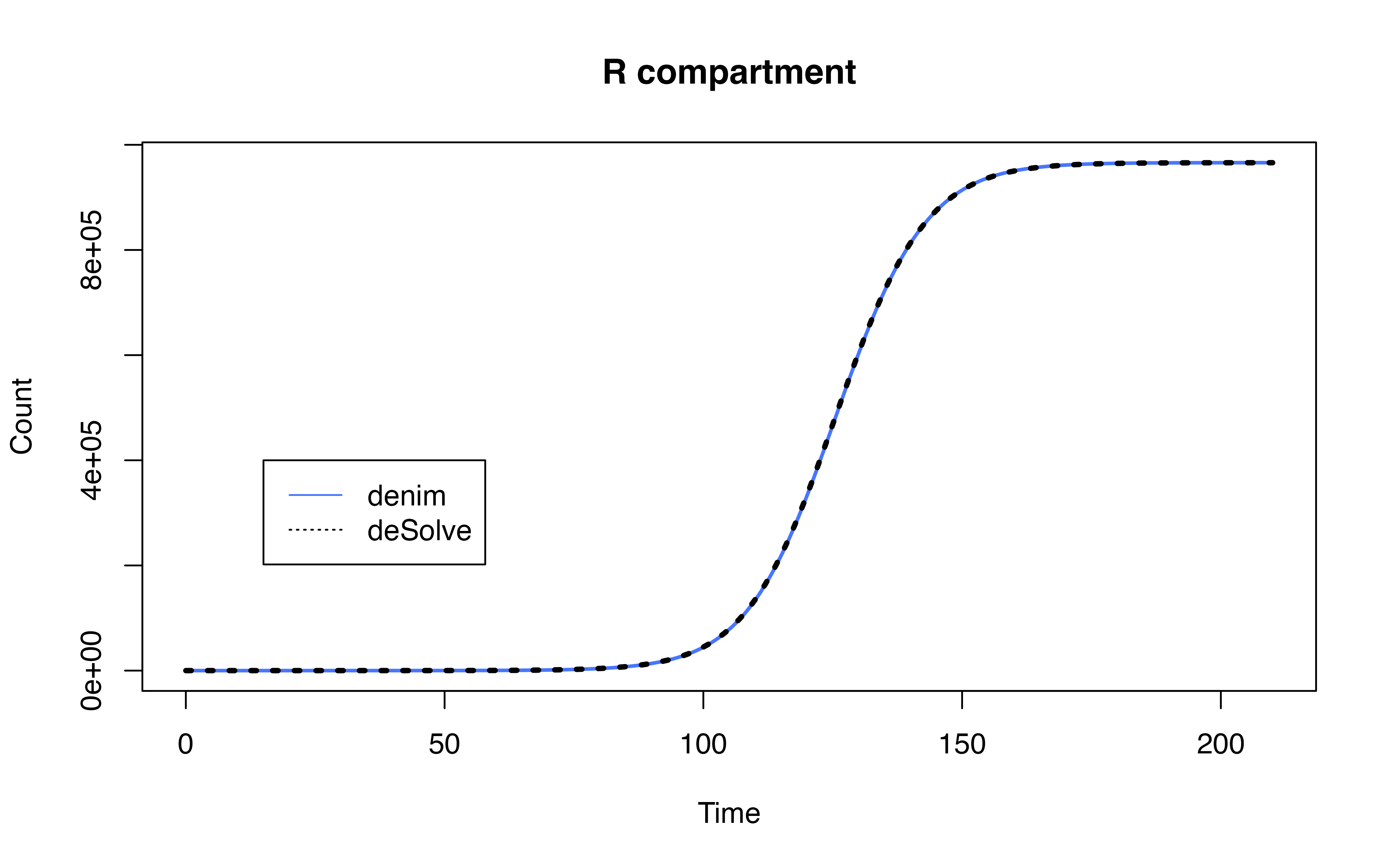
Comparison with SEIR model
Implementation using deSolve
model definition deSolve
library(deSolve)
parameters <- c(gamma_rate_I = 1/4, shape_I=2,
gamma_rate_R = 1/3, shape_R = 2,
R0 = 3.5, N = 1e6)
initialValues <- c(S = 999999, E1 = 1,
E2 = 0, E = 0, I1=0,
I2=0, I=0, R=0
)
# --- Transition def for deSolve
transition_func <- function(t, state, param){
with(as.list( c(state, param) ), {
tr = (1/gamma_rate_R)*shape_R
dS = - (R0/tr) * S * I/N
# apply linear chain trick
dE1 = (R0/tr) * S * I/N - gamma_rate_I*E1
dE2 = gamma_rate_I*E1 - gamma_rate_I*E2
dE = dE1 + dE2
dI1 = gamma_rate_I*E2 - gamma_rate_R*I1
dI2 = gamma_rate_R*I1 - gamma_rate_R*I2
dI = dI1 + dI2
dR = gamma_rate_R*I2
list(c(dS, dE1, dE2, dE, dI1, dI2, dI, dR))
})
}
times <- seq(0, 210, 1)
ode_mod <- ode(y = initialValues, times = times, parms = parameters, func = transition_func)
# --- show output
ode_mod <- as.data.frame(ode_mod)Implementation using denim
model definition in denim
denim_model <- list(
"S -> E" = "(R0/tr) * timeStep * S * (I/N)", # formulate according that of uSEIR method
"E -> I" = d_gamma(rate = 1/4, shape = 2),
"I -> R" = d_gamma(rate = 1/3, shape = 2)
)
initialValues <- c(S = 999999, E = 1, I= 0, R= 0)
parameters <- c(R0 = 3.5,
tr = 3*2, # compute mean recovery time, for gamma it's scale*shape
N = 1e6, timeStepDur = 0.01)
mod <- sim(transitions = denim_model,
initialValues = initialValues,
parameters = parameters,
simulationDuration = 210, timeStep = 0.01)
# denim_out[, c("S","E", "I", "R")] <- denim_out[, c("S","E", "I", "R")]/1e6
plot(mod)