install.packages(c("lubridate", "ggplot2", "epitrix"))
install.packages("epiparameter", repos = c("https://epiverse-trace.r-universe.dev", "https://cloud.r-project.org"))11 Generation interval
Install the following packages.
We will use a fictional Ebola outbreak linelist from The Epidemiologist R Handbook.
11.1 Definition
Definition 11.1 Generation interval is the time from the infection of a primary case to infection of the cases he/she generates (Cori et al., 2013).
If we match this to an SEIR model, the generation interval starts when the infector enters the E compartment (meaning they got infected). The latest time they can infect an infectee is at the end of their residence in the I compartment (they are infectious). Therefore, generation interval is the sum of residence times in compartments E and I (Gostic et al., 2020).
The mean generation interval decreases when susceptible persons are at risk of infectious contact from multiple sources. Because when a susceptible person has multiple potential infectors, there is a “race” to infect him/her in which only the first infectious contact leads to infection (Kenah et al., 2008).
11.2 Serial interval
Definition 11.2 Serial interval is the time between symptom onset in an infector–infectee pair (Gostic et al., 2020).
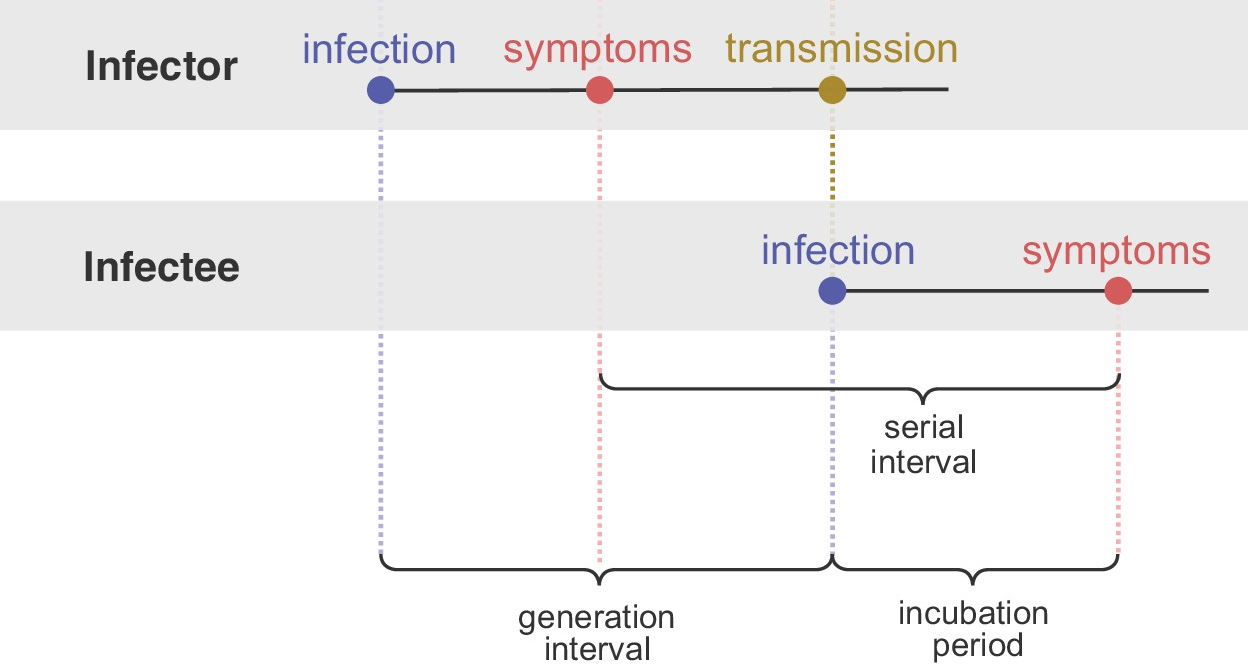
Serial interval is more easily observed than generation interval and often used in its place. Although the serial and generation intervals are often conflated, there are important differences (Gostic et al., 2020):
- Serial interval and generation interval usually have the same mean, but different variances.
- Serial interval can be negative (when the symptom onset of infector is later than infectee, such as in COVID-19), generation interval is always positive.
The distributions of the serial interval and the generation interval are identical in these scenarios (Cori et al., 2013):
- For diseases such as influenza, SARS, measles, and smallpox, it is expected that infectiousness starts only around the time of symptom onset.
- When the infectiousness profile after symptoms is independent of the incubation period.
11.3 Estimating serial interval distribution
library(lubridate)
library(ggplot2)
library(epitrix)Read and clean the data
df <- read.csv("data/ebola_si.csv")
# Convert date_onset to date type
df$date_onset <- dmy(df$date_onset)
head(df)In this data, we know who infected whom, and the date of symptom onset. To estimate the serial intervals, we need the time between symptom onset of each infector–infectee pair. Now let transform the original data to something like this:
| Infectee | Infector | Infectee’s date of onset | Infector’s date of onset | Time between onset |
|---|
# Get pairs of infector-infectee
tmp <- data.frame(infectee = df$case_id, infector = df$infector, infectee_onset = df$date_onset)
# Now let find the infector's date of onset
onset <- data.frame(case_id = df$case_id, date_onset = df$date_onset)
tmp <- merge(tmp, onset, by.x = "infector", by.y = "case_id", all.x = T)
colnames(tmp)[4] <- "infector_onset"
# Compute the time between onset
tmp$si <- as.numeric(difftime(tmp$infectee_onset, tmp$infector_onset, unit = "days"))
# Remove NA rows
tmp <- tmp[!is.na(tmp$si),]
head(tmp)We use function fit_disc_gamma() from package epitrix to fit a discretised gamma distribution to these serial intervals.
si_fit <- fit_disc_gamma(tmp$si)
si_fit$mu
[1] 11.51047
$cv
[1] 0.6686137
$sd
[1] 7.696056
$ll
[1] -6524.11
$converged
[1] TRUE
$distribution
A discrete distribution
name: gamma
parameters:
shape: 2.23691478711637
scale: 5.14568870945413We can plot this gamma distribution to compare with the observed serial intervals data.
si_distr <- data.frame(x = 0:max(tmp$si), y = dgamma(0:max(tmp$si), shape = si_fit$distribution$parameters$shape, scale = si_fit$distribution$parameters$scale))
# Scale the density to compare with serial intervals histogram
si_distr$y <- 4300 * si_distr$y
ggplot(tmp, aes(x = si)) +
geom_histogram(bins = 50, color = "white", fill = "blue", alpha = 0.4) +
geom_line(data = si_distr, aes(x = x, y = y)) +
labs(x = "Serial intervals", y = NULL) +
theme_minimal()11.4 Searching for serial interval distribution
library(epiparameter)Step 1. Load the database.
epi_dist_db <- epidist_db()Returning 122 results that match the criteria (99 are parameterised).
Use subset to filter by entry variables or single_epidist to return a single entry.
To retrieve the citation for each use the 'get_citation' functionStep 2. To see a full list of the diseases and distributions, use the parameter_tbl() function.
dt <- parameter_tbl(multi_epidist = epi_dist_db)
dtStep 3. Get the parameter of the disease you want.
ebola_si <- epidist_db(
disease = "Ebola",
epi_dist = "serial interval"
)Returning 4 results that match the criteria (4 are parameterised).
Use subset to filter by entry variables or single_epidist to return a single entry.
To retrieve the citation for each use the 'get_citation' functionebola_si# List of 4 <epidist> objects
Number of diseases: 1
❯ Ebola Virus Disease
Number of epi distributions: 1
❯ serial interval
[[1]]
Disease: Ebola Virus Disease
Pathogen: Ebola Virus
Epi Distribution: serial interval
Study: WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
Epidemic after One Year — Slowing but Not Yet under Control." _The New
England Journal of Medicine_. doi:10.1056/NEJMc1414992
<https://doi.org/10.1056/NEJMc1414992>.
Distribution: gamma
Parameters:
shape: 2.188
scale: 6.490
[[2]]
Disease: Ebola Virus Disease
Pathogen: Ebola Virus
Epi Distribution: serial interval
Study: WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
Epidemic after One Year — Slowing but Not Yet under Control." _The New
England Journal of Medicine_. doi:10.1056/NEJMc1414992
<https://doi.org/10.1056/NEJMc1414992>.
Distribution: gamma
Parameters:
shape: 4.903
scale: 3.161
[[3]]
Disease: Ebola Virus Disease
Pathogen: Ebola Virus
Epi Distribution: serial interval
Study: WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
Epidemic after One Year — Slowing but Not Yet under Control." _The New
England Journal of Medicine_. doi:10.1056/NEJMc1414992
<https://doi.org/10.1056/NEJMc1414992>.
Distribution: gamma
Parameters:
shape: 2.068
scale: 7.301
[[4]]
Disease: Ebola Virus Disease
Pathogen: Ebola Virus
Epi Distribution: serial interval
Study: WHO Ebola Response Team, Agua-Agum J, Ariyarajah A, Aylward B, Blake I,
Brennan R, Cori A, Donnelly C, Dorigatti I, Dye C, Eckmanns T, Ferguson
N, Formenty P, Fraser C, Garcia E, Garske T, Hinsley W, Holmes D,
Hugonnet S, Iyengar S, Jombart T, Krishnan R, Meijers S, Mills H,
Mohamed Y, Nedjati-Gilani G, Newton E, Nouvellet P, Pelletier L,
Perkins D, Riley S, Sagrado M, Schnitzler J, Schumacher D, Shah A, Van
Kerkhove M, Varsaneux O, Kannangarage N (2015). "West African Ebola
Epidemic after One Year — Slowing but Not Yet under Control." _The New
England Journal of Medicine_. doi:10.1056/NEJMc1414992
<https://doi.org/10.1056/NEJMc1414992>.
Distribution: gamma
Parameters:
shape: 1.898
scale: 6.532
# ℹ Use `parameter_tbl()` to see a summary table of the parameters.
# ℹ Explore database online at: https://epiverse-trace.github.io/epiparameter/articles/database.html